AlphaFold 3 Advances the Future AI Technologies for Pharma and Biotech
Bringing you the highlights on AlphaFold 3, we discuss how despite its restricted use for non-commercial applications, it augurs a bright future for fundamental biology, pharma and biotech, for example by paving the way to novel methods for virtual screening.
You know at Nexco we are always on the lookout for groundbreaking innovations in bioinformatics, in order to keep you informed and ourselves updated with the latest technologies that can help you in your projects in the best possible way. Today, we are thrilled to share the latest leap forward in structural biology: AlphaFold 3 [1]. Read on to know more about this model, its implications, its limitations, and also to see it in action with what we pose is the basis for the future of drug discovery.
Developed by DeepMind and Isomorphic Labs, both from the Google giant, AlphaFold 3 is not just another AI model for protein structure prediction… it’s probably the best tool for this as of today, and most importantly, it is one of the three only AI models that can parse not just proteins but also nucleic acids, ions, and ligands (the other being RoseTTAFold and the more experimental Umol, that we covered here from their preprints). This capability to understand multiple types of molecules and to integrate them upon modeling has far-reaching implications not only for fundamental biology but also for practical applications in the pharmaceutical and biotech sciences and industries, as we discuss next.
Revolutionizing Molecular Docking
One of the most exciting applications of AlphaFold 3 lies in using it to work as a molecular docking engine; one where you don’t need any kind of starting molecular structures to dock. Rather, the docking happens concurrently with the 3D modeling of the molecules (either proteins, nucleic acids, or small molecule ligands) as tertiary structure prediction happens concomitantly with modeling of the higher-order structure (akin to how AlphaFold 2-multimer modeled protein complexes by “cofolding” the proteins involved).
The possibility of using AlphaFold 3 to model complexes contrasts strongly with traditional methods for docking, which begin with input protein structures that are allowed to change very little (if at all) while only the internal degrees of freedom of small molecules are sampled. With AlphaFold 3 (and with its only other full counterpart from the academic world, RoseTTAFold-AllAtoms) researchers might in principle model the complexes between small molecules and protein targets in a radically novel way more tolerant to conformational changes relevant for intermolecular binding. We pose that this could revolutionize how we design and test new drugs, paving the way for faster and more cost-effective treatments. And the Google companies probably think so, that’s why they have filed patents, not released any code or weights, and only made the system accessible through a website with a limited number of submissions per day per Google account.
Notice that we speak in the above paragraph with terms like “might” and “could”. This is because although the technology is obvious very promising, right now the set of small molecules that AlphaFold 3 can handle is very limited. Therefore, you can’t really input your own small molecules and dock them to the protein, let alone a large library as you would do in a virtual screening campaign. However, our point is that the technology seems to be reaching the maturity required for this type of in silico analyses.
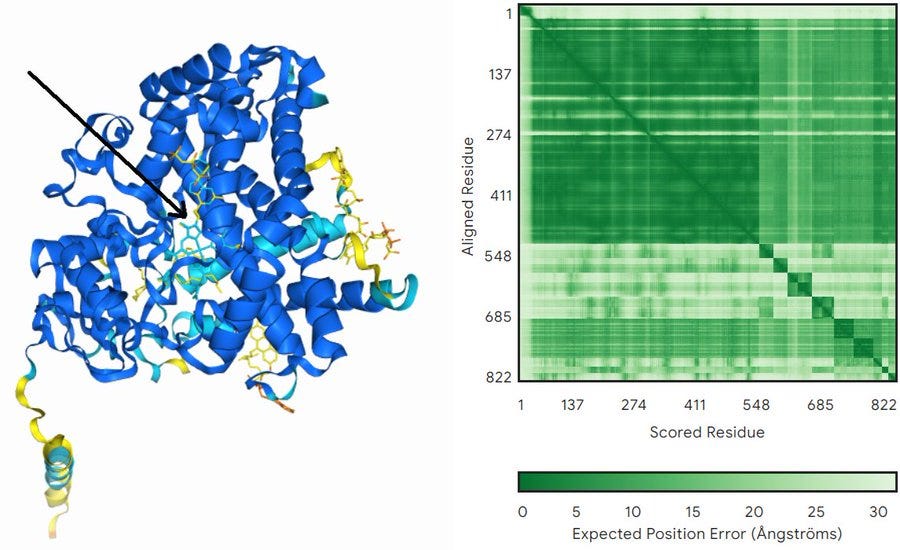
To further make our point, we will now share with you our own test on using AlphaFold 3 for molecular docking and some primitive form of virtual screening (figure below). We gave AlphaFold 3 the sequence of a xanthotoxin hydroxylase of unknown structure plus all the ligands available in the model, and after around 3 minutes we got back a pretty good model for the structure of the protein with all ligands bound in multiple places. Of them, all but one were assigned low confidence (yellow in the figure)… The only ligand modeled with high confidence is the heme group, which is the one actually expected to work at the core of this class of enzymes. Just this simple test would take hours to setup and run with regular docking/virtual screening program, with the additional disadvantage that you would need to model the protein before running the docking protocols themselves.
A Glimpse into the Future
While the use of AlphaFold 3 is at the moment restricted to non-commercial applications, we have witnessed in the recent past how academics and private companies built on the knowledge drown from AlphaFold 2 to build their own AI models with paralleled performance, for example OpenFold and RoseTTAFold. Even AlphaFold 3 itself has a competitor, already for some time, from the academic world, namely RoseTTAFold-AllAtoms, which might have produced less of a hype yet it is as promising as AlphaFold 3.
No doubt that these tools and similar ones to come have a huge potential to transform every aspect of the drug development process, the most obvious one being how we do molecular docking and virtual screening, as exemplified above with the toy example.
AlphaFold 3 and Nexco
At Nexco, we are always dedicated to helping our clients harness the full potential of the tools that come out one after the other, helping to solve their problems and answer their questions better and faster. While AlphaFold 3 cannot be used for commercial applications, our team has the expertise to leverage AlphaFold 3 for non-commercial purposes, such as academic research. From exploring molecular interactions to predicting protein structures, we’re here to support our clients every step of the way.
More broadly, as we embrace this new era of innovation with tools like AlphaFold 3 and RoseTTAFold-AllAtoms and everything that will for sure come next, we are committed to staying at the forefront of structural bioinformatics and molecular modeling so that we can provide our clients with the tools and expertise they need to succeed.
References
[1] https://www.nature.com/articles/s41586-024-07487-w
Related Posts

Nos locaux:
Nexco Analytics Bâtiment Alanine Route de la Corniche 5B 1066 Epalinges, Switzerland
Appelez-nous
+41 76 509 73 73
Laissez-nous un message
contact@nexco.chN'hésitez pas à nous laisser un message
Nous nous ferons un plaisir de vous répondre et de trouver des solutions optimales à vos besoins.
