ChIP-seq
ChIP-seq, a foundation for epigenomics
Epigenomics has acquired a preponderant role in the study of genomics. Complementary to the study of gene expression through transcriptomics techniques such as RNA-seq or scRNA-seq, the inquiry of the chromatin status at the loci of interest has become crucial to understanding gene expression. Chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) is a fundamental technique in epigenomics for which we have years of experience.
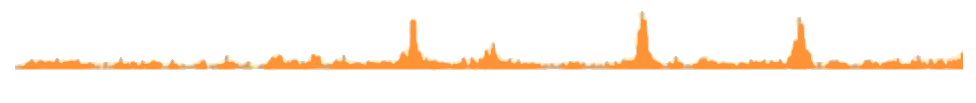
Reveal protein binding sites and discover DNA motifs
ChIP-seq allows to study the binding pattern of your protein of interest through the entirety of the genome. Nexco analytics method to analyze ChIP-seq and infer binding sites (peaks) is in accordance with current standards. Our pipeline first focuses on quality control to discard any bias and to minimize false negatives by utilizing advances statistics.
Motif discovery is usually the following step in the analysis. Find out the DNA sequence recruiting your protein of interest in your data by taking advantage of our motif detection pipeline.
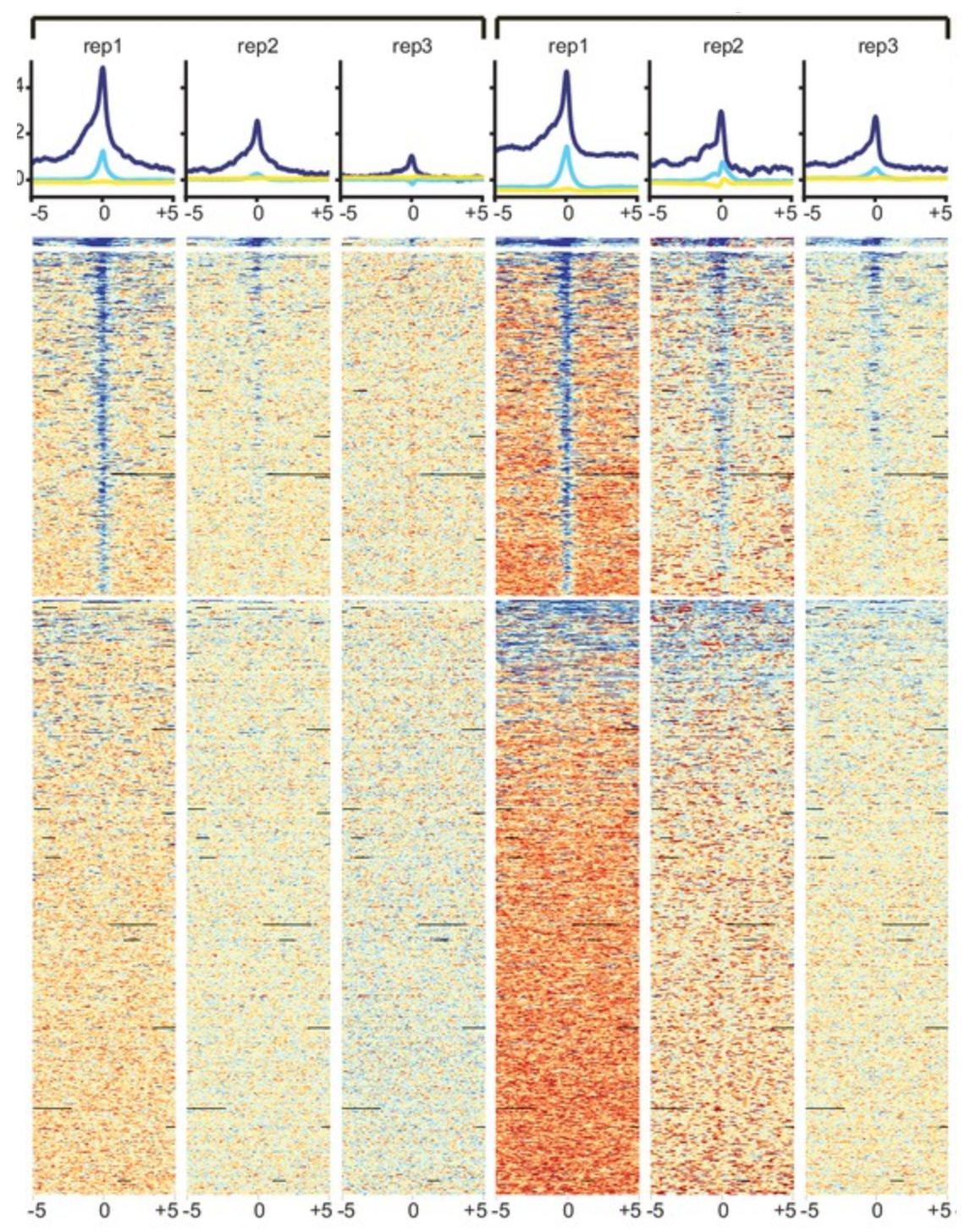
Explore chromatin status via histone marks
By targeting histone marks, ChIP-seq allows to study the chromatin landscape across the genome. Discover in your data which regions are open, closed, enhancers, promoters and couple it with transcriptomics to get a better understanding of gene expression. Nexco analytics' multidisciplinary team has been working in epigenomics for a decade and are experts in histone marks.
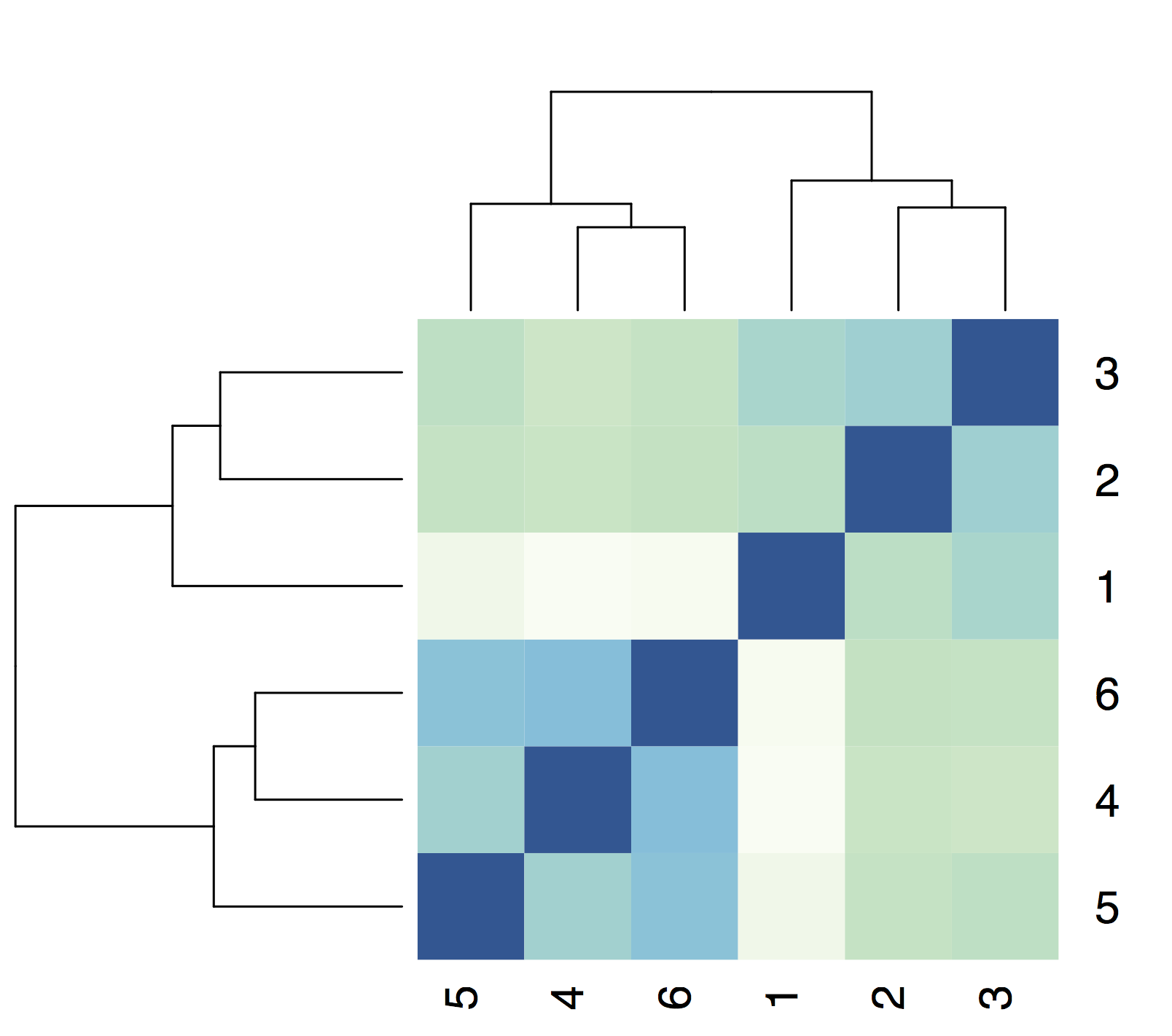
Cross with public databases
As epigenomics became more prevalent, consortiums started the systematic generation of ChIP-seq data targeting known transcription factors and histone marks in common cell lines. Those public data form a mine of information that we can dig for you. By crossing your data with public datasets, discover if your protein of interest DNA binding correlates with other transcription factors or histone marks in different biological background.
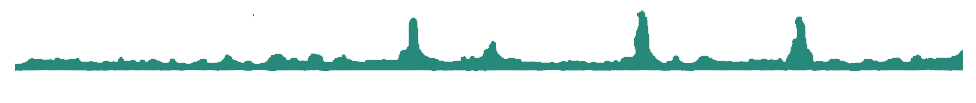
Multi-omics
Gene expression has to be looked at through the prism of epigenomics. By crossing your transcriptomics data with ChIP-seq data, it is possible to assess the regulatory role of your protein of interest by correlating the binding to the expression of the targeted genes.
Further analyses can then be run on the putative targets, like pathways analysis, gene ontology or others to characterize the targets and find biological relevant results.
