scRNA-seq
scRNA-seq: transcriptomics at single cell level
Single-cell RNA-seq is becoming the new standard in transcriptomics. It allows the analysis of the full transcriptome at single cell resolution. This relatively new technique is rapidly evolving and we are keeping our pipelines up to date with the constant advances in the field.
Expertise in all single cell flavors
Many different approaches exist when it comes to scRNA-seq. Some allow a good sensitivity per cell at the expense of a reduced number of cells as it is typically the case for SMART-seq3. Others, like 10x allows to assess the transcriptome of thousands of cells but at a lower sensitivity.
Nexco Analytics has experience in all the different subtypes of scRNA-seq and we provide the adequate and optimized pipeline for each.
Cell type labelling, clustering and classification
One of the powers of scRNA-seq is to allow the classification and labelling of cells in your sample. Nexco Analytics is using published methods to extract this information and provide you with digested results ready for interpretations.
After cell clustering, it is possible to infer the gene markers that categorize the specific cell types. Get insights on which genes are crucial in your experiment and discover new cell subtypes.
Go an additional step by computing the trajectory of cells in your samples, allowing you to observe cell changes through time and trace back their history.
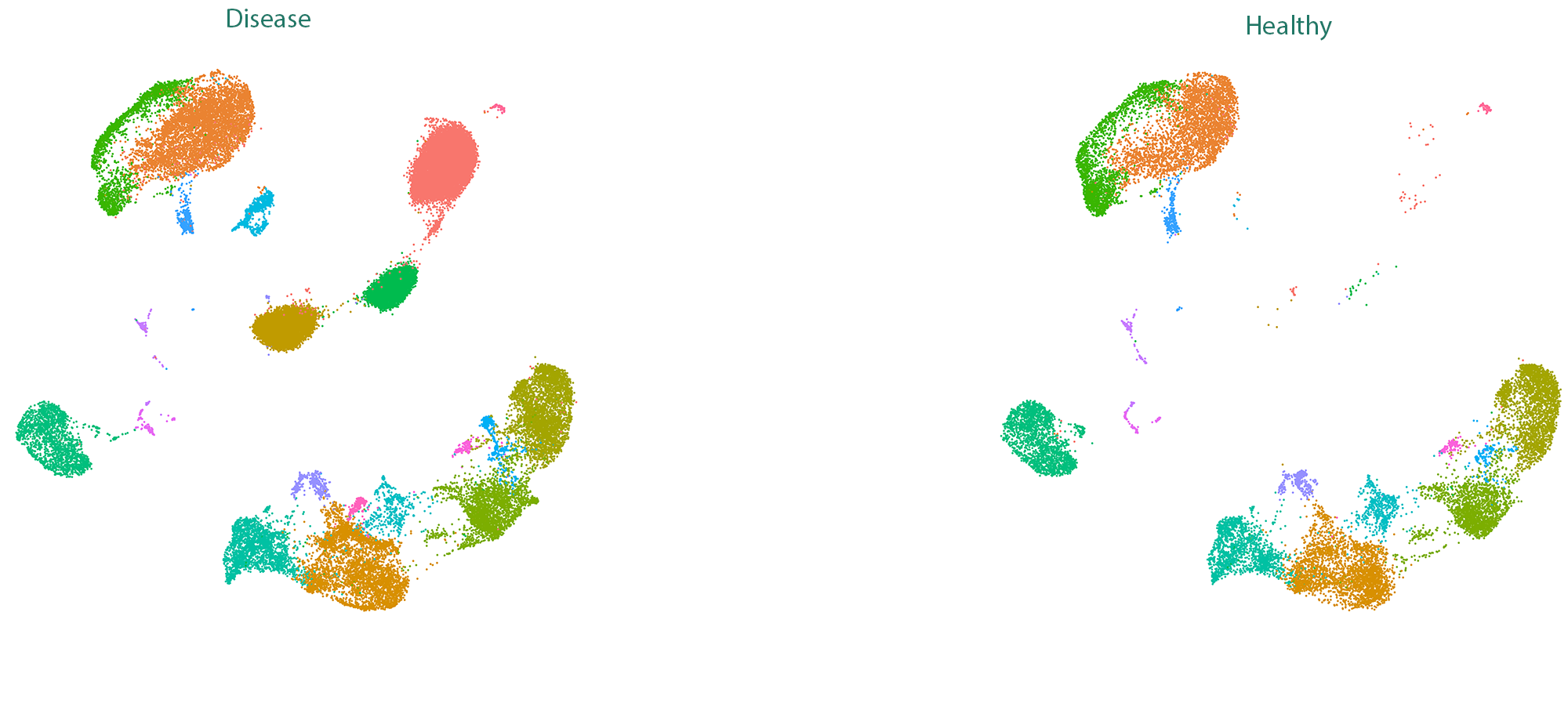
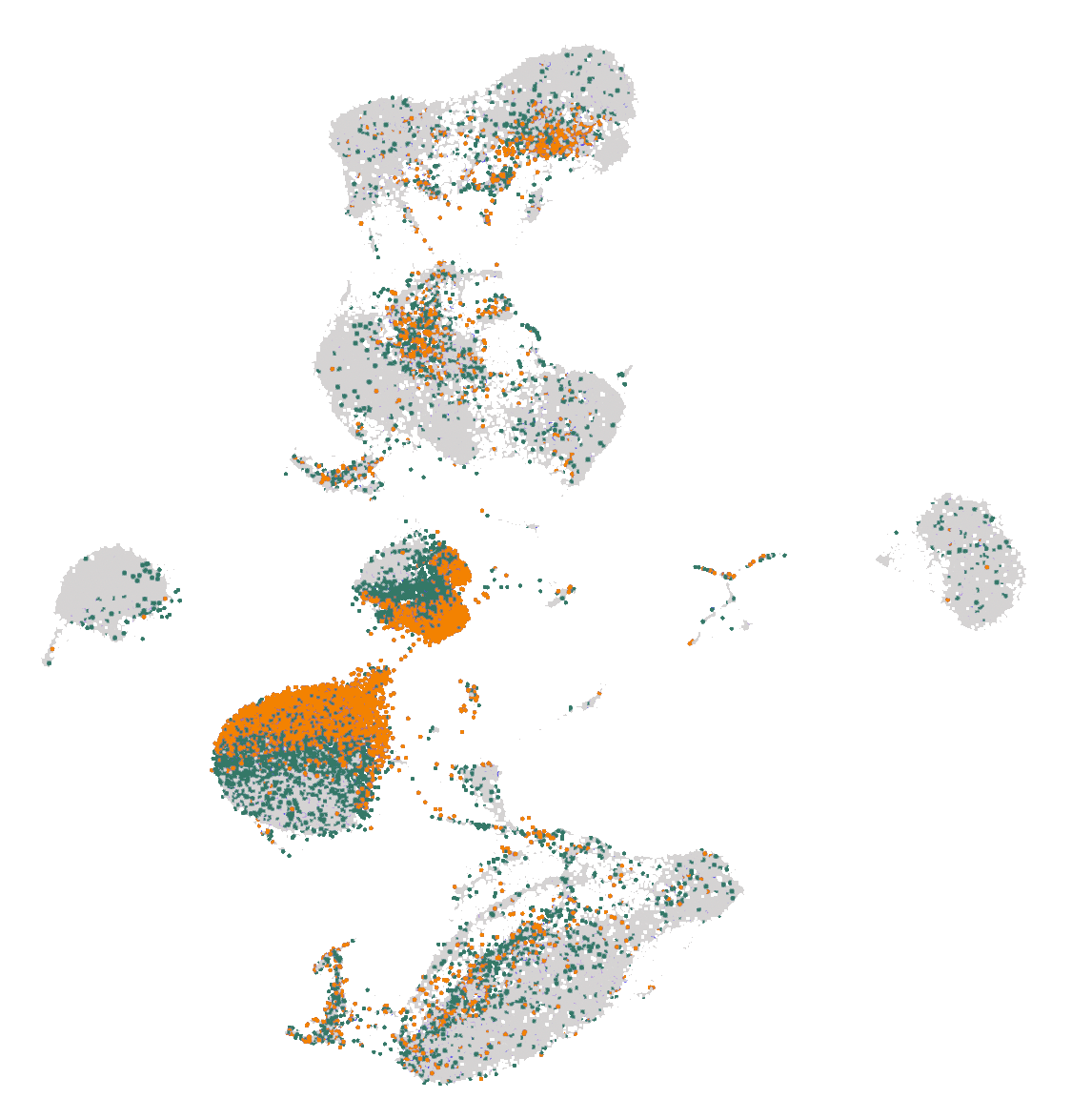
Transposable element quantification at single-cell level
Nexco Analytics TEnex pipeline allows to study transposable elements in scRNAseq data. Our method has been developed at EPFL and licensed to Nexco Analytics and can cope with the general lower sensitivity of scRNA-seq compared to bulk RNA-seq. Find out how transposable elements contribute to your experiment and make the most of your data using a higher level of granularity than the one provided only by genes.
The next level with Multi-omics and spatial transcriptomics
Recently, scRNA-seq protocols have been improved to interrogate the chromatin status of the cells at the same time as the gene expression pattern. This technique, combining ATAC-seq and scRNA-seq, narrows the results by reducing false positives in the expression data. Even if you are not using those recent methods, we can cross your data with other techniques to fit your needs.
Another technique that is growing is spatial transcriptomics, where cells are barcoded according to their spatial localization in tissues. Thanks to our experience, we can apply even the most advanced techniques and help you interpret your results.

