2024 at Nexco, Growing and Innovating in the Era of -Omics and Artificial Intelligence
Spatial transcriptomics, multimodal AI systems like AlphaFold 3, large language models, and immersive 3D graphics systems that are here to stay — this year’s highlights are all in this post

Throughout 2024 we at Nexco have undergone remarkable growth and achievements, following from close and participating at the frontiers of computational biotechnology, and including you too in our vision. We will keep pushing the boundaries of bioinformatics, harnessing the power of artificial intelligence (AI) and cutting-edge technologies to accelerate research and drive innovation in the life sciences. This dedication to advancement is clearly reflected in our blog posts from the past year, showcasing our commitment to staying ahead in all these rapidly evolving fields that affect fundamental biology, applied biotechnology, and the pharma.
In this last post for 2024 we cover four pivotal technologies that we have extensively covered in our blog due to their major relevance:
- Spatially resolved -omics, especially spatial transcriptomics, with which we can interrogate the molecular landscape of tissues in high detail and at high resolution in 2D and 3D.
- Multimodal AI agents that promise to model biology holistically, spanning from genomics to higher omics and structural biology, thuis promising a revolution for structural biology and drug development.
- Powerful large language models that can assist work in bioinformatics and virtually any kind of computer work in ways we didn’t even imagine just a decade ago.
- Immersive yet affordable hardware and software for augmented, virtual and mixed realities, that became finally practical, affordable, and useful, promising to revolutionize how we explore and discuss spatial data, molecular structures, and more in 3D.
We will also show you how we engaged with the sector by assisting to the most important events in biotech, hiring new talents, and developing and testing novel software and tools to stay at the forefront of the field.
Riding the Wave of -Omics: Spatial Transcriptomics Takes Center Stage
Nexco’s core expertise lies in dissecting complex biological data. Its mastery of -omics analyses, particularly spatial transcriptomics, shines brightly. This revolutionary technique, that we covered in detail here and here with examples here, allows researchers to visualize the activity of genes directly within their tissue environments. This means they can literally view in 2D or 3D, by using software like Open-ST covered here, the activities happening inside each cell or even inside different compartments of a cell or tissue:

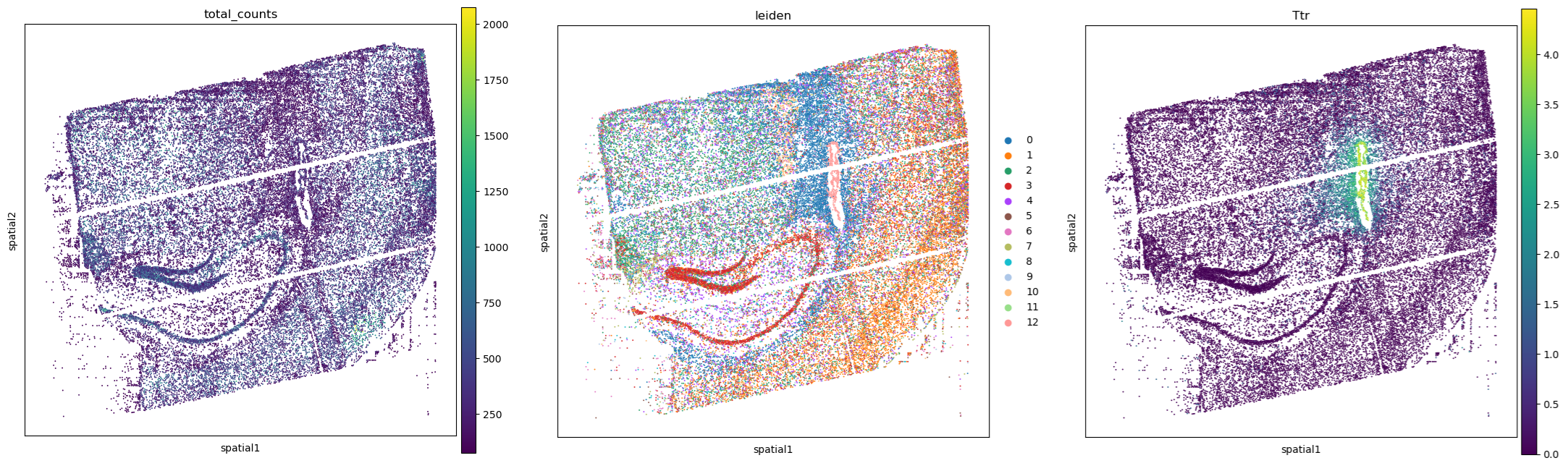
Nexco’s prowess in analyzing spatial transcriptomic data besides just bulk single-cell RNA-seq, and also our several other pipelines ready to process data from various -omics, empowers researchers to unlock a deeper understanding of biological processes and to find novel disease markers and treatments.
Recognizing the need for robust and reliable data processing, we provide streamlined, data-secure workflows ensuring accurate and biologically meaningful results. From deciphering positional barcodes and mRNA sequences to identifying cell-cell interactions and spatial trajectories within tissues, Nexco helps researchers with all aspects of spatial transcriptomics data analysis.
And right now we are introducing a novel software, ONex, that allows customers to directly interrogate -omics data themselves, even without being experts and without any need for coding skills.
Democratizing Access to -Omics Analysis with our ONex Software
At Nexco we understand that the advancements in Next-Generation Sequencing (NGS) technologies have led to a surge in data volume, creating a demand for streamlined and standardized analysis tools for all the -omics that involve sequencing. To meet this need, we are developing ONex, a platform designed to simplify and enhance the -omics data analysis process.
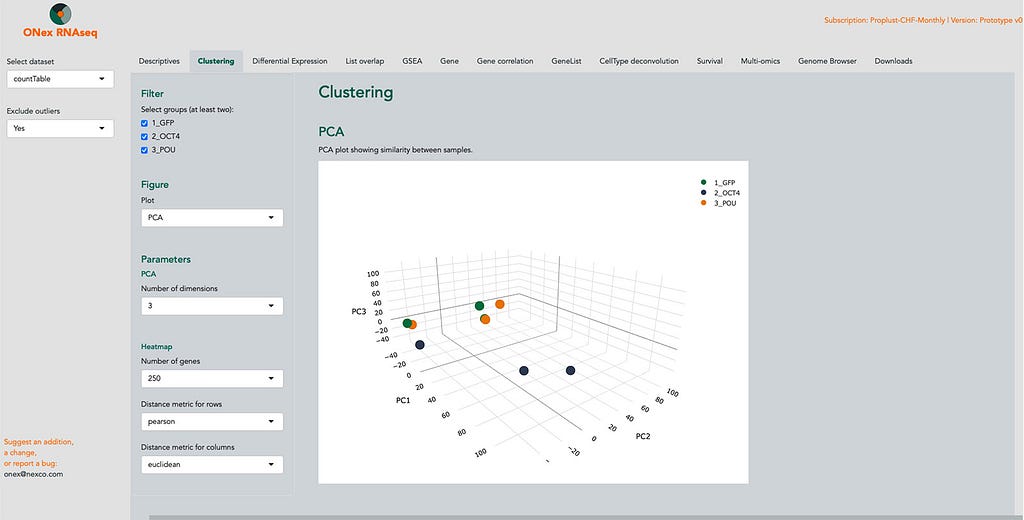
Currently being tested with real-world users, ONex offers several very important advantages:
- Comprehensive and standardized analysis: ONex features optimized pipelines for various -omics applications, currently including transcriptomics and soon epigenomics, genomics, and proteomics.
- Accessibility and ease of use: Users can access and run analyses on ONex directly from their web browsers, removing the complexities associated with software installs, updates, parametrization, etc.
- Data security: ONex is hosted on servers located in Switzerland, known for its stringent data privacy laws. In addition, it can be installed on-premises when dealing with sensitive data.
AI-Powered Structure Prediction: From AlphaFold 3 to New Frontiers in Drug Design
The 2024 Nobel Prize in Chemistry underscored the transformative impact of AI on protein structure prediction. We at Nexco embrace these advancements, recognizing the game-changing potential of tools like AlphaFold 3 from two Nobel Prize winners working at Google Deepmind and also of more recent, less restrictive alternatives like Chai, Boltz, RoseTTAFold-AllAtoms from the 2024 Nobel Prize D. Baker, and others. As we covered here for AlphaFold 3, these AI models are capable of not just predicting protein structures but also modeling their interactions with other molecules, including drugs. This groundbreaking capability is revolutionizing drug design by enabling researchers to simulate and analyze protein-ligand complexes like never before.
Traditional methods for docking small molecules into protein targets are often limited by conformational changes. These new multimodal AI systems tackle this challenge head-on, paving the way for faster, more efficient, and cost-effective drug development strategies. See for example how easily you can model a protein-drug complex with full conformational freedom using Chai Discovery’s program, just to try one case where we provide as input the sequence of the TEM lactamase together with that of a substrate, nitrocefin:
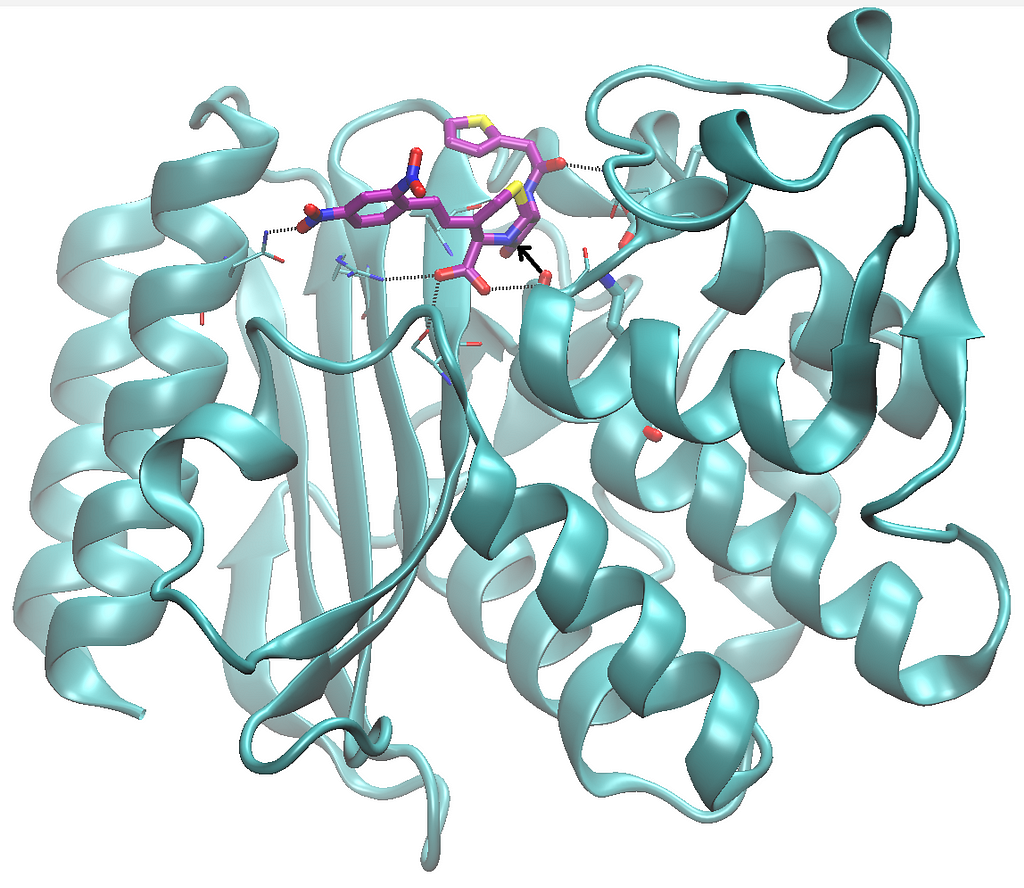
Most notably from this 2024, the various breakthroughs in protein structure prediction meant that scientists can now do many other kinds of also very important predictions. First of all, they can now better predict the effects of mutations on proteins, as we covered with Deepmind’s AlphaMissense. And probably most impactful so far, they can now better predict sequences that fold in 3D as intended, that is they can design proteins ad hoc, as we also covered in our blog including also example applications to stabilizing enzyme for biotechnology.
Multimodal, “Holistic” AI systems Spanning from Genomes to Transcriptomes and Biomolecular Structures
The above-described multimodal models are only “structural”, treating biological molecules and their complexes atom by atom just as you need to compose 3D models of their structures starting from protein and nucleic acid sequences. But as we have also covered in our blog, multimodal systems that span genomes, transcriptomes and proteomes all as “strings” from which information emerges, are also being created. Some of these models, such as gLM2 trained from the Open Meta Genomics corpus (preprint here), have been show to capture coevolution information that reflects protein-protein interactions. Some protein sequence-only models such as ESM from Meta can be tuned to predict protein structures and have also been shown to have learnt about protein biophysics and evolution upon training.
In the view of our experts, the ground is set for a new leap whereby scientists can connect, in single huge AI models, the world of letters in genomics and transcriptomics with the world of atoms and chemistry relevant to structural biology and drug design. When this will be achieved as a single huge multimodal model that can grasp everything from genes to transcriptomes, proteomes, perhaps metabolons, and to the chemistry and physics underlying molecular function, we will have unlocked a totally new way to do biology, biotechnology, and medicine.
Large Language Models Transforming Bioinformatics
In our blog posts and in our work, we have also recognized the power of large language models (LLMs) in streamlining bioinformatics workflows, semi-automating data analysis, assisting with report write up and text mining, and more.
It is obvious that LLMs, much like the technology behind ChatGPT, are revolutionizing how researchers approach data analysis and knowledge extraction. But we specifically know how to actively integrate these LLMs into our software and pipelines, harnessing their capabilities through programmatic means for tasks such as text mining and ontology, querying unformatted text and data, building smart assistants and agents, automatizing data annotation, labeling datasets, and more.
One particularly interesting application of LLMs in which we and others are actively working is in the development of natural language interfaces that enable researchers to interact with their data and software using simple, intuitive language commands in natural language, even through natural speech.
For more about LLMs and some concrete example applications, check out our introduction to LLMs for biology and bioinformatics, our post on how to maximize the power of bioinformatics with LLMs (also here), and a summary about how companies and academics are innovating with LLMs for research and development.
Augmented, Virtual, and Mixed Realities Come of Age: Powerful and Accessible, Now Here to Stay
There have been already a few waves of hype around augmented reality (AR), virtual reality (VR), and mixed reality (MR), but we are positive that the one we are experiencing now is the good one, and is here to stay. With billions invested not only by giants like Meta but also by other big names like Apple as well as tens of small companies bringing very exciting developments and ideas, we are sure that the technology has finally reached a level of maturity that makes it poised for widespread adoption.
AR, VR and MR technologies are no longer just futuristic novelties or experimental tools. They have become affordable, compact, truly wearable, accessible, and remarkably intuitive to use. At Nexco, we are confident that this new generation of immersive experiences will stay and profoundly impact how we visualize and interact with data, particularly in the life sciences.
Immersive 3D visualization technologies are uniquely suited for tackling one of the major challenges in modern science: the understanding of complex data in 3D, leaving behind the limitations imposed by 2D screens. Although software development is growing steadily but somewhat slowly, we are positive that the technology‘s capabilities are powerful enough to allow, today, the exploration of intricate systems in 3D space. Modern devices can even track hands and limbs, thus allowing users to interact with virtual objects with their bare hands, as we have shown in some examples by connecting with our neighbors at EPFL who specialize in the development of immersive multiuser interfaces for molecular graphics and modeling.
Revolutionizing Multiuser, Interactive Molecular Graphics and Modeling
A particularly exciting application of AR, VR and MR technologies is in interactive molecular graphics and modeling, which seem otherwise stuck with flat screens, mouse and keyboard operations, and other highly unnatural means to interact and visualize molecules.
Immersive environments let scientists wander freely inside a world where they can manipulate virtual representations of molecules — of any type, including proteins, DNA, materials, etc. — in natural, very intuitive ways. This enhances 3D understanding by allowing researchers (and students and teachers, because these tools have been proven very useful in education) to examine biomolecular interactions and dynamics in ways that are just impossible to achieve with traditional 2D interfaces.
Here is, as an example, the complex between TEM-1 lactamase and nitrocefin derived above with Chai discovery’s AlphaFold 3-like server, as seen through MolecularWebXR, a free multiuser AR/VR tool for 3D presentations and discussions in chemistry and biology:
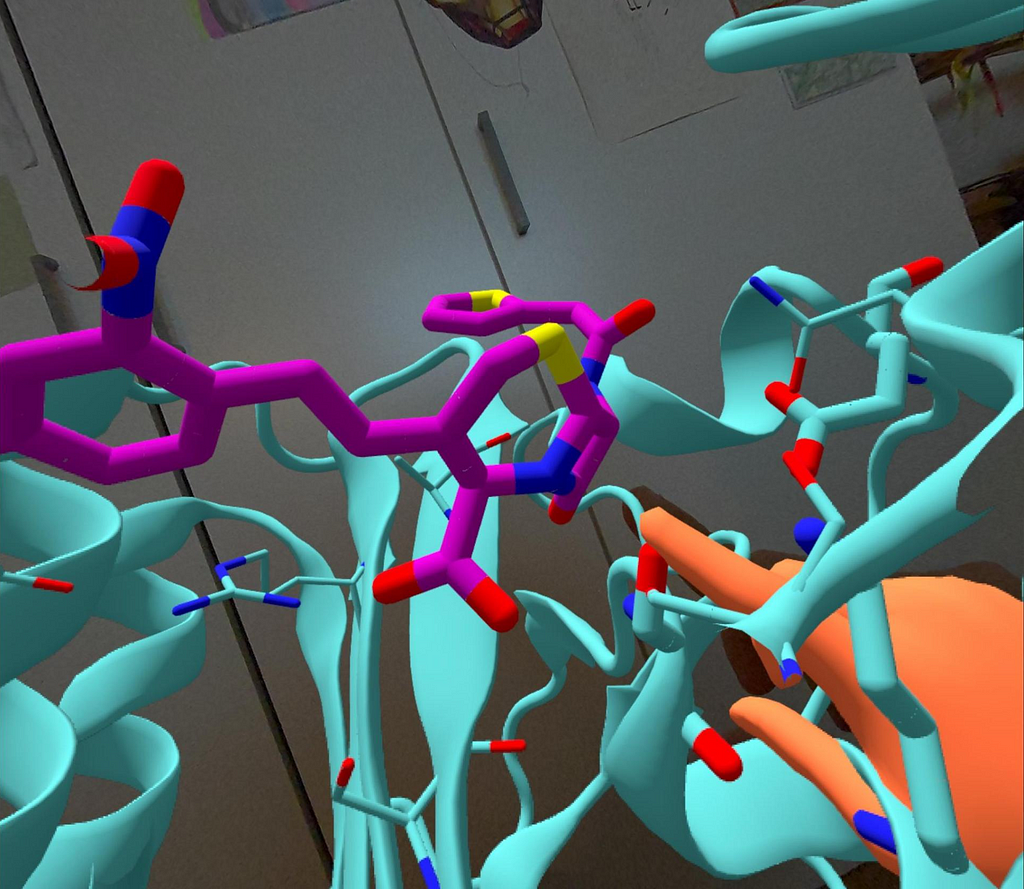
Other modern tools, also from our next-door colleagues at EPFL, go a step further and allow not just immersive multiuser visualization but also acting on molecules through interactive molecular simulations. Their prototype, called HandMol, allows users to manipulate molecular structures with their hands, leveraging cutting-edge technologies to create a seamless and very natural experience that makes operations with keyboard and mouse look so obsolete!
By combining WebXR for immersive visualization, exquisite hand tracking, AI-assisted molecular mechanics, speech recognition powered by large language models, and web-mediated connections for multiuser collaboration, HandMol stands as a true pioneer in modern molecular graphics and modeling. This integration of advanced technologies means that users can naturally interact with molecular structures in 3D space while the computer handles a realistic simulation of the underlying physics and chemistry.
Wearing a VR or AR headset, users can then grab, move, reshape, build, break and deform molecules in a virtual environment as if they were physical objects.
The platform’s speech recognition system, coupled to an LLM from OpenAI, allows users to control features and perform actions using natural language commands, further lowering the barrier to entry and enhancing usability. The collaborative capabilities in turn allow multiple users to share the same virtual space, interact in real time with the same molecular system, and combine efforts for tasks like drug design, protein engineering, and molecular education.
As we embrace 2025 and thanks to privileged connection with these colleagues at EPFL, Nexco will continue exploring how tools like HandMol can revolutionize data analysis, for and beyond molecular graphics and modeling as these technologies clearly have many other applications.
Nexco Connecting and Collaborating at Key Industry Events
Again in 2024, we actively participated in industry-leading events to connect with peers, showcase innovations, and foster collaborations. Our presence at the Swiss Biotech Day 2024 highlighted our commitment to driving innovation and empowering users with transformative solutions.

During the two Swiss Biotech Days we proudly took the chance to present our software ONex to prospective clients, in a booth extended with hands-on demonstrations of our next-door developers of immersive molecular graphics.
This year we also had a place at the European edition of BioTechX, a conference that brings together companies working in genomics, translational medicine, data science, IT, bioinformatics, cheminformatics, healthcare and pharma. We again took the chance to showcase our innovations while presenting success cases and our software at the startup poster session. It was also the opportunity to listen to interesting talks about the future of AI in precision medicine.

Expanding Our Team
In 2024, our team experienced significant growth with the addition of three talented professionals. These new team members have primarily strengthened our sales and marketing efforts while also bringing valuable expertise to further enhance our bioinformatics capabilities. A warm welcome to them again!
Closing a Great 2024 for Bioinformatics and for AI Applied to the Life Sciences
With several new projects ongoing and growing through grants, hirings, and collaborations, and with our eyes on modern technologies as we have shared with you through our blog, we close a great 2024 and anticipate an exciting 2025 ahead. We hope to keep leading bioinformatics and AI for life sciences in a highly practical way, transforming how our customers plan, explore, execute, analyze and understand their experiments to get tangible and efficient advances in their research and development projects.
Related Posts

Our location
Nexco Analytics Bâtiment Alanine, Startlab Route de la Corniche 5A 1066 Epalinges, Switzerland
Give us a call
+41 76 509 73 73
Leave us a message
contact@nexco.chDo not hesitate to contact us
We will answer you shortly with the optimal solution to answer your needs
